Bridge RNAs are here to take CRISPR's crown
Move over CRISPR, bridge RNAs are here to make the precision genome edits that you can't.
A lot of noise has been made about genome engineering in the last two decades.
But the idea of engineering genomes really began in the 1980’s with the discovery of mammalian, viral, and bacterial recombination systems.
In the early days we envisioned evolving these tools to precisely manipulate large pieces of DNA within living cells.
We quickly realized that was very hard and were only really able to do it reliably in embryos or in plant seeds.
Large scale engineering in living organisms would have to wait.
But then along came RNA interference, TALENs and their friend CRISPR which allowed us to do some of those cool things we had initially imagined, just at a smaller scale.
RNAi allowed us to chop up RNA’s we didn’t want around and prevent the expression of specific genes.
TALENs and CRISPR allowed us to make single nucleotide edits or create small insertions and deletions in specific genes.
Some of us even attempted to use CRISPR to insert new sequences into specific places in the genome.
But that process is very messy, highly inefficient and really only works in situations where you can take cells out of someone’s body, edit them, make sure those edits are correct, and then put them back in.
Wouldn’t it be nice if you could just take a big piece of DNA that you want in a new place and use an enzyme to efficiently stick it in there without all of this extra fuss?
We might be in luck.
Because a new genome editing system has been identified in bacteria that uses a small guide RNA and a recombinase to specifically insert a donor DNA sequence into a target.
And the entire system is available in a tiny package that should be slightly easier to get into cells than the other competing alternatives.
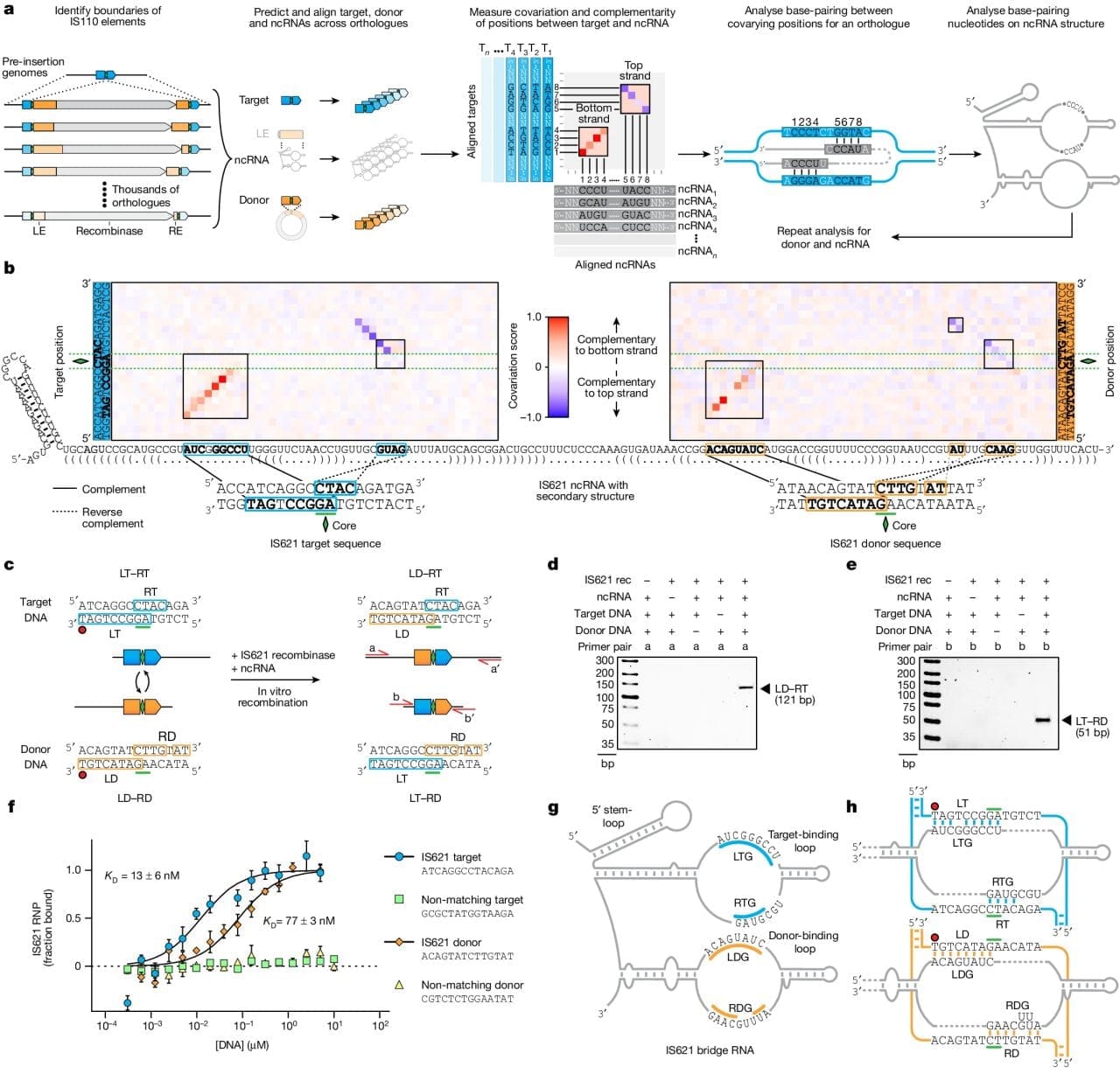
The figure above shows how it was discovered and how it works. a) bioinformatic identification of guide RNA sequences found in IS110 recombinase family members. b) heat map identifying two sets of highly conserved regions. c,d,e) this system is able to perform a recombination reaction in a test tube. f) In the presence of its guide RNA, IS621 (an IS110 family member) has very high affinity for target and donor sequences. g) a graphical representation of how IS621 uses its guide RNA to ‘bridge’ the donor and target DNA together.
The authors went on to show that this system is programmable and can make specific edits.
And, interestingly, because the IS110 family is so large, there are a number of recombinase/guide RNA combinations to explore that might provide additional functionality or increased edit specificity.
Bridge RNAs look like a promising new tool in our genome engineering arsenal that might someday allow us to make the edits we've always dreamed of.

