De novo mutations are more common than we previously thought
There’s more to de novo mutations (DNMs) than just SNPs and indels.
Human genetic variation, or the differences in our genomes that contribute to why each of us look, sound, and behave differently, are the result of de novo mutations.
But, before my genetics friends start screaming at me, I just mean that that variation had to be de novo at some point in the past!
Because the textbook definition of a DNM is:
“A mutation or alteration in the genome of an individual organism that was not inherited from its parents.”
DNMs are important because they are the mutations that serve as the basis for the generation of new disease phenotypes, new protein functions and ultimately the evolution of a species.
Because it’s these new mutations that increase or decrease an organism’s ‘fitness’ or ability to survive long enough to produce offspring.
But how do we know if a mutation is de novo or just something that was passed down from one of our parents?
With sequencing!
By looking at how these mutations arise and are transmitted between generations, we can calculate a rate of their occurrence.
We’ve tried to do this before, but because the dominant sequencing technology for the past two decades has been short-reads, we’ve missed important classes of DNMs, which in turn has caused us to underestimate the DNM rate!
These missed mutations include structural variants (SVs), variants in highly repetitive sequences, and variants in things like pseudogenes and paralogs.
To help get a more accurate rate calculation, researchers have now sequenced the CEPH 1463 family, a well-documented 28-member, four-generation pedigree using both short and long read sequencing technologies to generate near-complete, phased assemblies (>95%) of each family member’s genome.
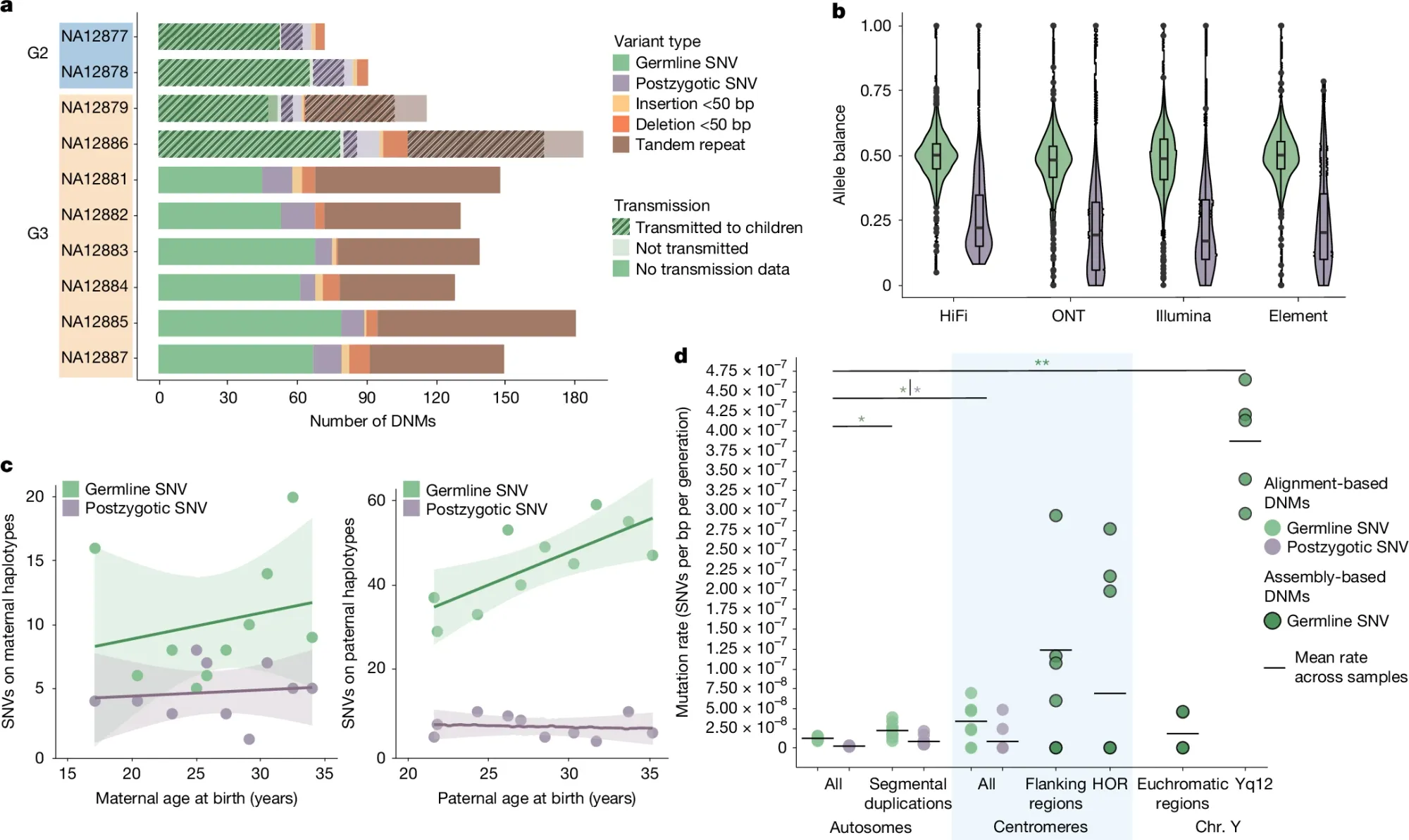
Part of what they found can be seen in the figure above which focuses on SNP and indel mutations: a) displays the different variant classes found across each generation, b) shows the allele balance of germline variants (green) vs post zygotic mutations (purple - happen after fertilization during development) c) Highlights that these mutations occur more often with parental age and that fathers are responsible for significantly more of them than mothers and d) that there’s significant enrichment of these events in the repetitive regions of chromosomes near the centromeres.
The paper goes on to calculate DNM rates for tandem repeats, SVs, and Y chromosome variants.
The researchers “estimate a range of 98–206 DNMs per transmission (average of 152 per generation) and observe a strong paternal de novo bias (70–80%) and an increase with advancing paternal age, not only for SNVs but also for indels and SVs, including TRs.”
This is an increase from the previously calculated DNM rate of 60-70 per generation and this work underscores how improvements to genome sequencing technology can help us to better understand the complexities of human genetic variation.
