Fuzzy Sequencing: Sometimes close enough is good enough
DNA is coded by 4 bases, but no one said that's how we have to sequence it. Ready to have your mind bent?
My brain was never more broken than when I tried to learn about SOLiD's color space but "Fuzzy Sequencing" is a close contender!
If you remember the ABI SOLiD sequencer, it operated VERY differently than the sequencing by synthesis technology that has come to dominate sequencing for the past 2 decades.
SOLiD was a "sequencing by ligation" technology that ligated fluorescently labeled nucleotide pairs together to determine the sequence of a target DNA molecule.
It also employed an incomprehensible color encoding scheme where 4 dyes were used to label the 16 (4x4) nucleotide pairs and the sequence was determined by deconvoluting the sequences obtained using 3 different sequencing primers (each shifted by one base).
If that sounds confusing, it is - but it helped introduce this concept that just because there's 4 bases, it doesn't mean you have to sequence them individually in a strictly linear fashion.
Oxford Nanopore (ONT) has done something similar with its sequence detection scheme where their pores aren't actually sequencing individual bases at any given time...they're predicting what Kmer (an ~ 6 base sequence) is present in the pore.
It's much easier to try to detect a Kmer than to detect the signal of a single base as it occludes a nanopore!
But both SOLiD and ONT use(d) their sequencing schemes to try to identify an accurate sequence of the individual bases contained in the DNA fragment being sequenced.
What if you didn't care what the sequence actually was?
Are there more efficient ways to identify a target molecule than by sequencing it at single base resolution?
Those might sound like insane questions, but some of the most lucrative applications of sequencing technology DO NOT require single base resolution to give us the answers we're seeking.
For example, most pathogen detection or microbial identification tests, non-invasive prenatal testing for chromosomal anomalies, or whole transcriptome profiling - basically anything where we're counting/detecting fragments - don't need single base resolution to accurately identify those things!
So, it could make sense to create a new sequencing technology that encodes genetic information more cost effectively and efficiently for those kinds of applications.
Enter Fuzzy Sequencing!
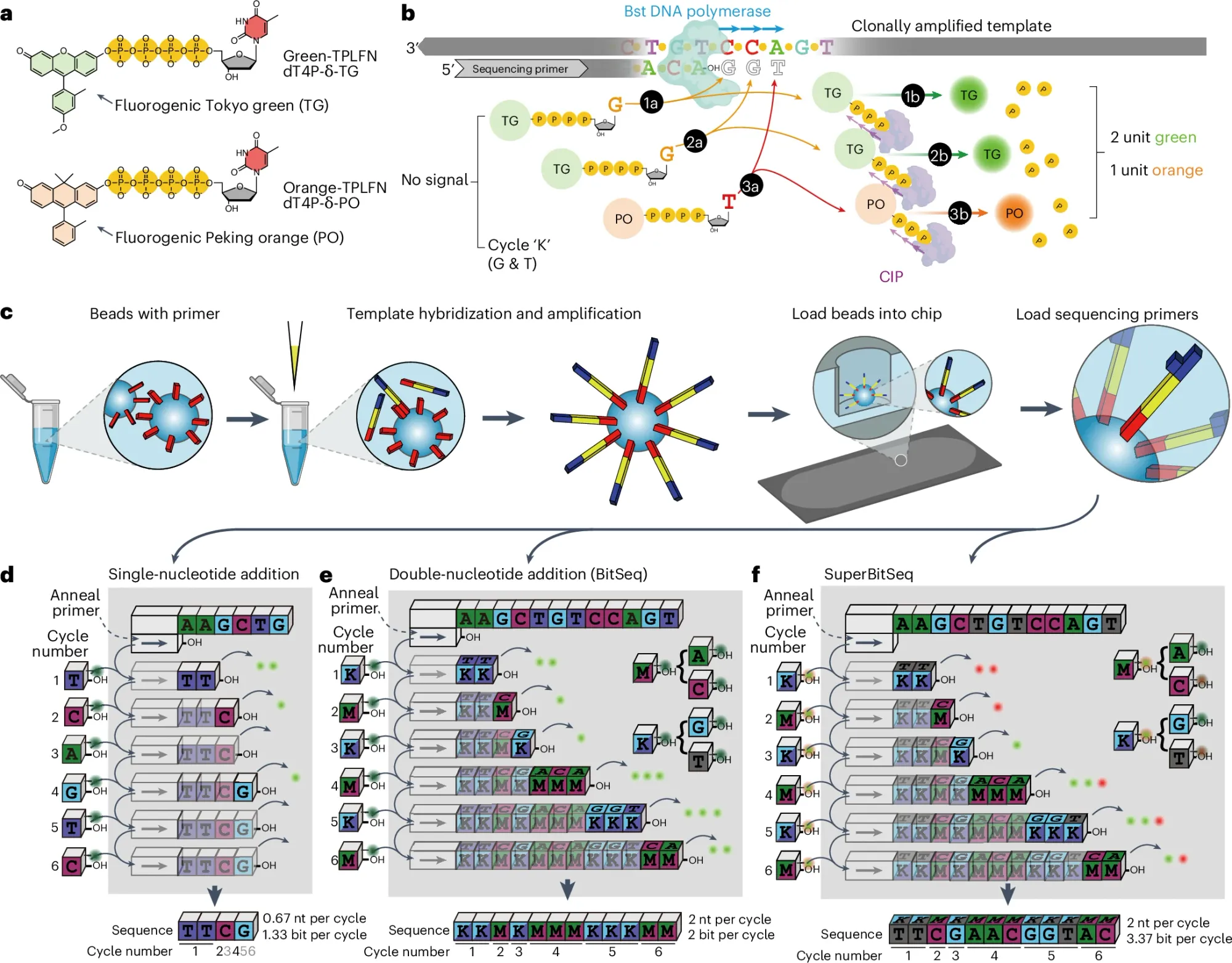
Instead of reading out individual bases, Fuzzy Sequencing provides a "Fuzzier" read out which is depicted in the figure above not as bases but as an encoded flowgram! (see d)
a) It uses one (BitSeq) or two (SuperBitSeq) fluorgenic dyes
b) tagged bases are added to each cycle as 'flowgrams' (eg K = G/T and M = A/C) are flowed together and unlike Illumina sequencing that uses reversible terminators, this chemistry does not, so multiple bases can be added every cycle (similar to how IonProton pyrosequencing works) but that's ok because the number of bases added can be determined by the fluorescence intensity of the signal.
c) the sequencing reaction is based on bead emulsion PCR where beads capture a single sequence, amplify it on the bead surface and then that bead is deposited in a nanowell for the sequencing reaction - here the sequencing cycles proceed by sealing and unsealing the wells with oil to prevent reagent diffusion or signal mixing
d) compares the sequencing efficiency of a single base addition per cycle to that of BitSeq and SuperBitSeq - which is to say that adding multiple bases per cycle is faster and cheaper with a readout of not A,C,T, and G but as K and M!
And if you're having trouble understanding how this encoding works to identify a genetic sequence, the researchers provided this fun diagram:
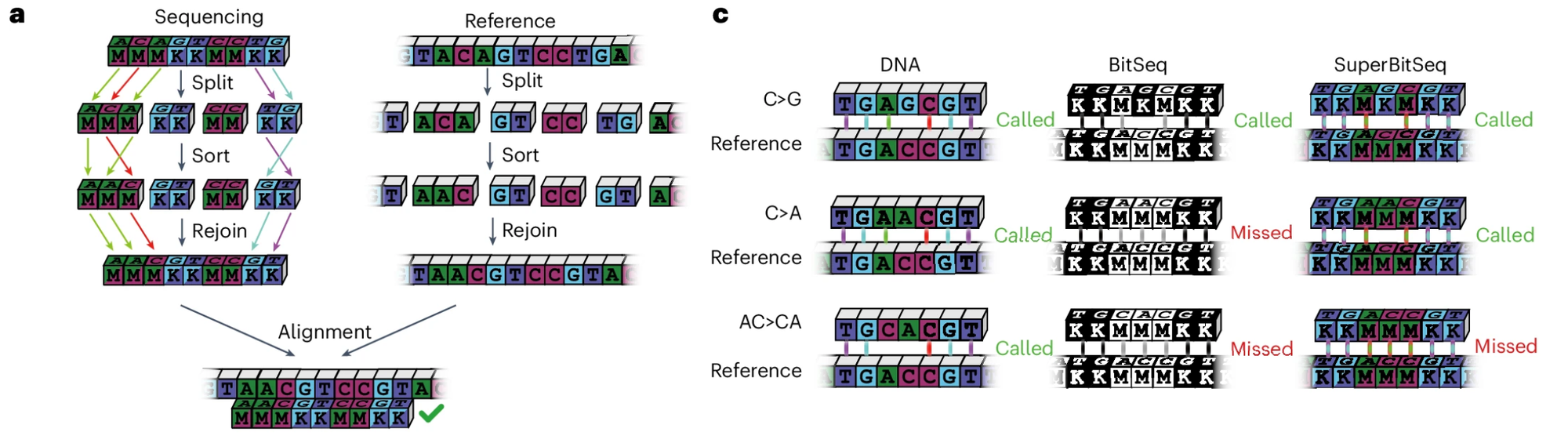
Obviously encoding information this way means that there will be base ambiguity at some positions (see 'missed' above), but for the applications where you'd be using this, you don't care!
The researchers show in follow-up experiments that Fuzzy Sequencing is able to accurately detect large copy number variations in prenatal testing cases (including chromosomal microdeletions) and identify microbes from throat and anal swabs.
While I don't see Fuzzy Sequencing taking the sequencing community by storm, it's an interesting thought experiment and if developed commercially could be a nice alternative to multiplex digital or real-time PCR.
It could also make prenatal testing, transcriptomics, and even oncology testing significantly more cost effective!
