In 2009, high throughput sequencing gave us our first glimpse of the 3-D organization of the DNA in our nuclei
While chromosomes are usually depicted as X's, they actually spend most of their time jumbled up like a giant ball of yarn.
Friedrich Miescher first discovered nuclein (DNA) in the early 19th century, and determined it was bound to proteins.
But it was Walther Flemming in 1889 who suggested that this DNA-protein complex be called chromatin - which is the material that makes up our chromosomes.
Don’t be fooled, though.
Just because chromatin was described and named in the 1800’s, that doesn’t mean anyone actually understood what it did!
It took until 1950 for scientists to finally begin to understand that the ‘genetic’ material was DNA.
And it wasn’t until the 1960’s that we first started piecing together how chromatin actually functioned.
To this day, we’re still not entirely sure!
But the classical depiction of our genetic material as ‘rods’ or ‘X’ structures in many introductory biology textbooks is not totally accurate.
This is because these structures only appear when chromosomes ‘condense’ during cell division!
The vast majority of the time, most of our chromosomes are unwound in the nucleus and tangled together in a giant mass.
You might be wondering if the interweaving of our chromosomes in the nucleus plays an important role in regulating which genes are expressed?
And the answer to that question is:
You bet your chromatids it does!
While it had been hypothesized for many years that long-range interactions played an important role in gene expression, it wasn’t until 2009 that these chromosomal contacts were first characterized genome-wide.
Previous work using a technique called chromosome conformation capture or 3C had shown that regions of chromatin could form ‘loops’ or touch each other at long distances.
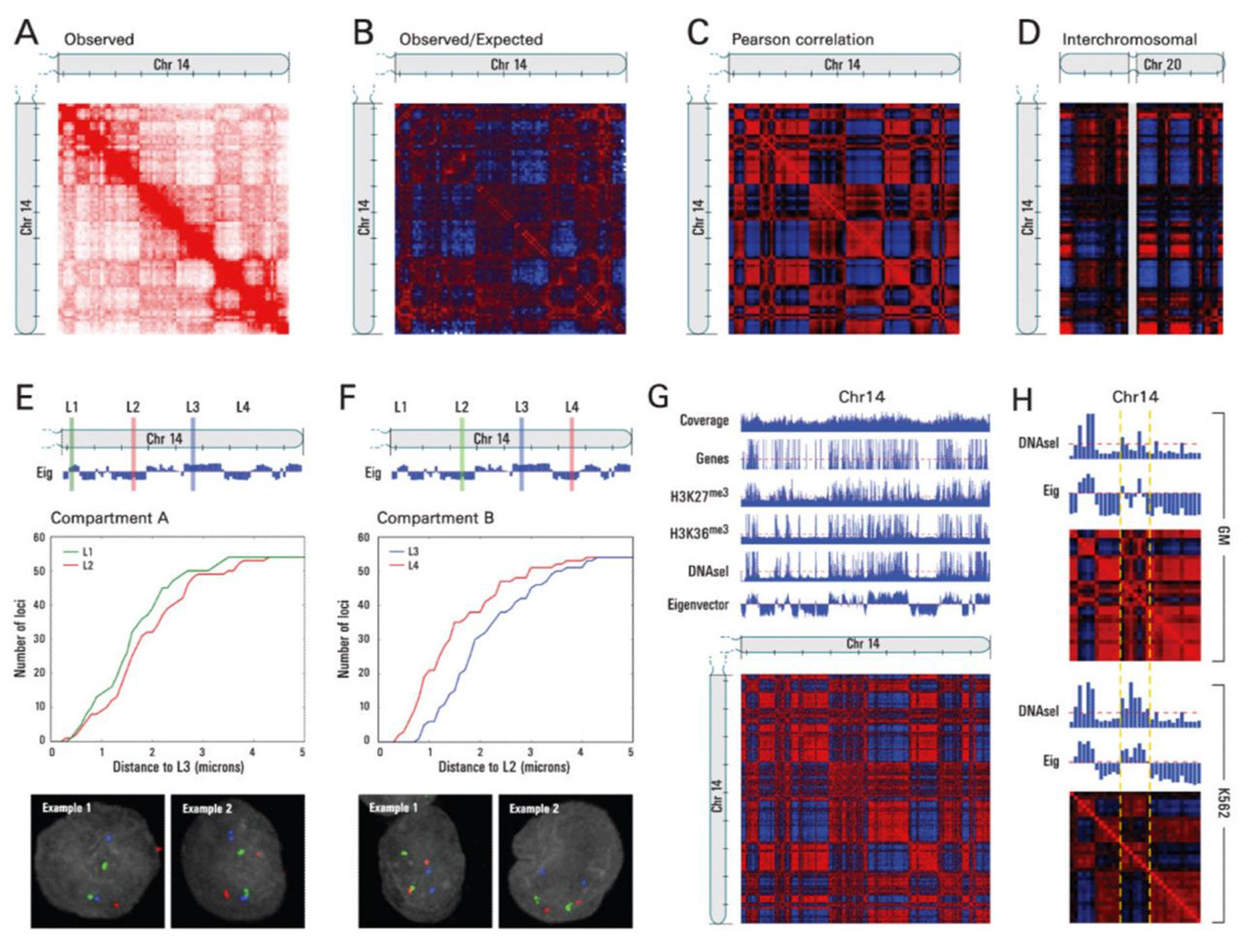
But, the authors here mapped chromosome contacts using a technique that they named Hi-C.
It differed from previous chromosome conformation capture methods in that it was the first to leverage high throughput sequencing to look at ALL of the connections within and between chromosomes to map the three-dimensional structure of the genome.
And what they found was really cool!
A) Shows a heat-map of interactions across chromosome 14, B) highlights areas of more contact than expected (red) and less contact than expected (blue), D) shows interchromosomal interactions between chromosome 14 and 20, E and F) highlight how linearly distant regions can actually be in very close proximity to one another, and G and H) show how this architecture correlates with gene rich, open and closed regions of chromosomes!
So, why are these results important?
Because they showed us how the physical three dimensional structure of our genome can impact gene expression, adding yet another regulatory layer to the story of epigenetics.

