A new genetic cause for Inflammatory Bowel Disease has been discovered
Inflammatory diseases affect 5% of the population but we have a poor understanding of how to treat them. Multi-omic studies are changing that.
Inflammatory and autoimmune diseases like lupus, psoriasis, Crohn’s disease and ulcerative colitis can be severely debilitating.
With respect to the last two, which are inflammatory bowel diseases (IBDs), symptoms can include rectal bleeding, severe abdominal pain, and diarrhea.
These conditions are usually diagnosed by looking at inflammatory markers in stool samples or viewing inflamed tissues via colonoscopy or endoscopy.
The exact cause of the inflammation in IBD is not well known, although flares in some people can be associated with eating habits or disruptions of the microbiome.
This has had the effect of confusing people about the difference between it and irritable bowel syndrome (IBS) which can have similar symptoms but whose cause is more related to infections (like C․diff), bacterial overgrowth, or other changes that affect gut motility.
But because IBD is associated with inflammation, and co-occurs frequently with other autoimmune diseases, it’s hypothesized that there is a strong genetic component to the disease.
Genome wide association studies (GWAS) have identified over 200 regions of the genome linked to IBD; however, only a few of these regions have mechanistic associations with inflammatory genes.
Unfortunately, many of these variants fall in non-coding regions or gene deserts and so it is thought (as is true for much of GWAS) that these associated regions are involved in epigenetic regulation of gene expression in affected cell types.
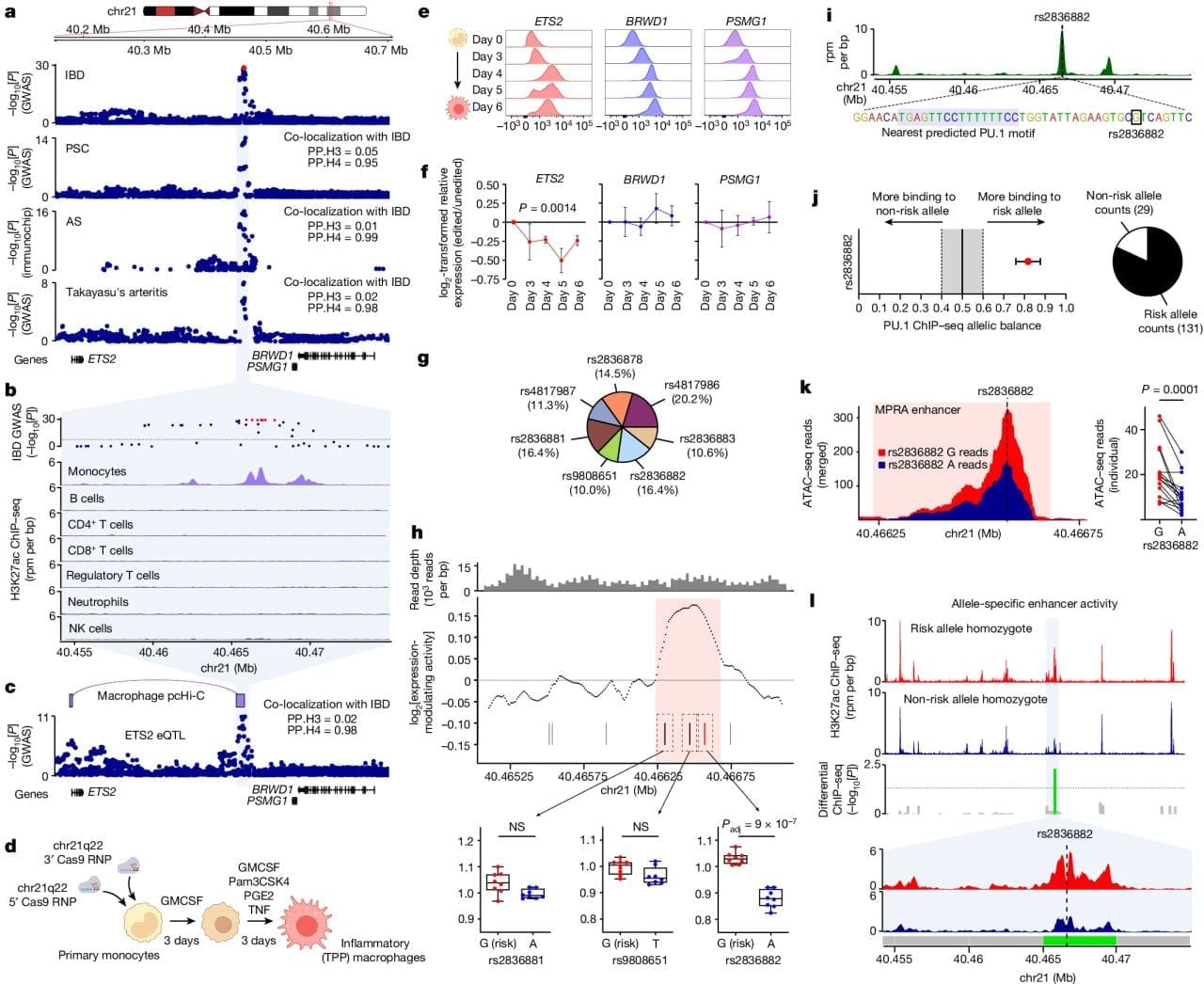
To that end, the researchers behind this week's paper chose to study a region of the genome associated with multiple inflammatory diseases, chromosome 21q22 (chr21q22).
In the figure above you can see a) the GWAS association of this region with IBD, primary sclerosing cholangitis (PSC), ankylosing spondylitis (AS) and Takayasu’s arteritis. b) this region is associated with activating H3K27ac epigenetic marks only in inflammatory monocytes. c) this region is physically linked to the ETS2 gene, a transcription factor. d, e, f) CRISPR removal of this region results in downregulation of ETS2 expression. g,h,i,j,k,l) a specific SNP, rs2836882, is associated with recruiting the chromatin remodeling protein PU.1 to this newly discovered ETS2 enhancer.
The authors go on to show that CRISPR–Cas9 disruption of ETS2 reduced pro-inflammatory markers and single-cell transcriptomics of diseased IBD tissue confirmed the presence of higher levels of ETS2 in associated monocytes.
While more work needs to be done, it appears that targeting the ETS2 pathway presents a promising therapeutic strategy for treating conditions like IBD.

