Omic.ly Weekly 78
June 9, 2025
Hey There!
Thanks for spending part of your week with Omic.ly!
This Week's Headlines
1) Real-Time monitoring of hospital pathogens pays dividends
2) Structural variant based MRD testing is coming to an oncologist near you
3) A Pile of Pennies
4) Weekly Reading List
Hospital genomic surveillance of pathogens stops outbreaks, prevents deaths and saves a bunch of money.
Hospital acquired infections (HAI) are a big problem in healthcare.
Not only are they expensive to treat, but they often lead to unnecessary deaths.
The most common pathogens involved in HAI's are Clostridioides difficile, Methicillin-resistant Staphylococcus aureus (MRSA), Vancomycin-resistant Enterococci (VRE), and multidrug-resistant Gram-negatives like Klebsiella pneumoniae and Pseudomonas aeruginosa.
The challenge in detecting these deadly bugs is that HAI monitoring has traditionally been a reactive exercise where we're always playing catch up when it comes to an infection or an outbreak.
Most hospitals rely on passive surveillance to detect a rise in infections above baseline levels which then leads to an investigation after an outbreak is already in full swing.
But the tools in our molecular testing toolkits have become cheaper and higher throughput now and could allow us to be more proactive about pathogen and outbreak surveillance.
One of the options here is to perform routine environmental monitoring of a hospital and its patients using whole genome sequencing.
This allows for early detection of outbreaks and enables quicker intervention.
However, widespread adoption of WGS for these activities in the hospital setting has been hindered by cost, lack of infrastructure, and limited institutional incentives.
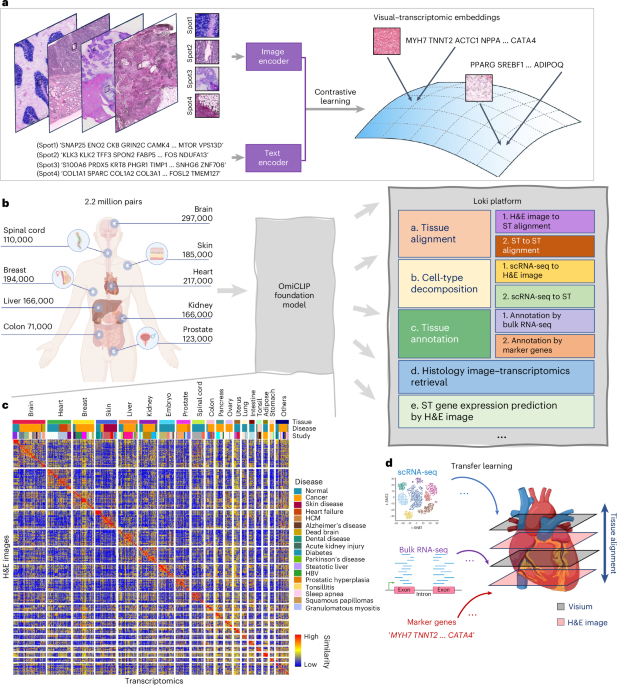
In response to these challenges, researchers developed the Enhanced Detection System for Healthcare-Associated Transmission (EDS-HAT).
This system integrates real-time WGS surveillance with algorithm-driven outbreak detection.
After a promising retrospective analysis of a Klebsiella outbreak in their endoscopy unit, the researchers then used the system in a two-year study that provided powerful evidence of its effectiveness:
1) They were able to detect outbreaks with as few as two positive cases
2) In 95.6% of outbreaks, interventions guided by WGS surveillance successfully halted transmission
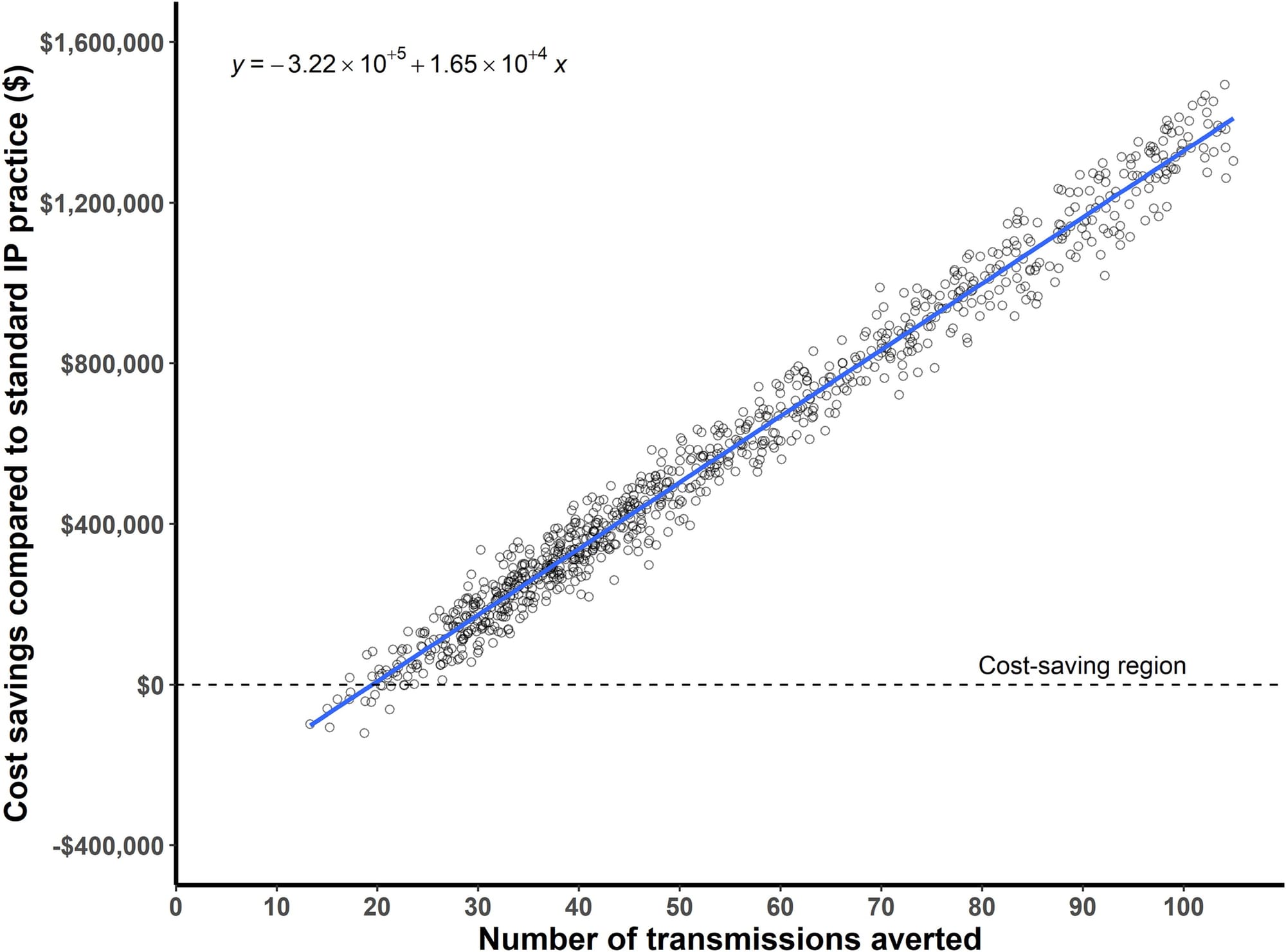
3) Real-time genomic surveillance had a net savings of $695,706, with a potential upper-bound savings reaching $1.77 million and 126 prevented infections (See figure above)
But most importantly, they estimate that 62 infections and 4.8 deaths were prevented during the study period.
This study offers powerful evidence that real-time genomic surveillance is feasible and highly effective in identifying and mitigating outbreaks.
So it goes without saying that integration of WGS-based surveillance seems like a no-brainer when it comes to hospital infection prevention and control practices.
###
Sunderman AJ, et al. 2025. Real-Time Genomic Surveillance for Enhanced Healthcare Outbreak Detection and Control: Clinical and Economic Impact. DOI: 10.1093/cid/ciaf216
Structural Variants: What are they good for? MRD testing, actually.
Minimal Residual Disease (the “M” can also stand for “Molecular” or “Measurable”) is a term used to describe the trace amount of cancer left in a patient’s body after treatment.
This is typically detected by tracking cancer specific biomarkers in blood over time to determine if or when a cancer recurs.
These tests are usually referred to as liquid biopsies!
But, back in the day, MRD tests were based on markers known to be common to specific cancer types.
Unfortunately, these early tests had limited value because cancer is cancer and not all of them behave predictably.
They’re all a little different!
They all contain mutations that are unique to them, and so if you’re not developing a test that’s specific to them, there’s a chance you might miss them if they do start to come back!
Thankfully, there are a couple ways around this problem.
One is to do very deep sequencing of blood every couple of months to find variants that appear to be related to a cancer, but doing a lot of deep whole genome sequencing can get pretty expensive!
That’s why a good number of MRD tests use a “tumor informed” approach where they sequence the tumor to find all of the variants in a sample and then pick a handful of those to create a “fingerprint” to track over time.
Creating a fingerprint of the tumor reduces cost and complexity while increasing sensitivity because now you actually know what you’re looking for!
But cancers can have a lot of different types of variants so it’s important to pick ones that you think happened early in development (because they’re probably really important for cancer survival!) and pick variants you can detect with high confidence.
For the longest time, the variants that were used in MRD were single nucleotide variants (SNVs).
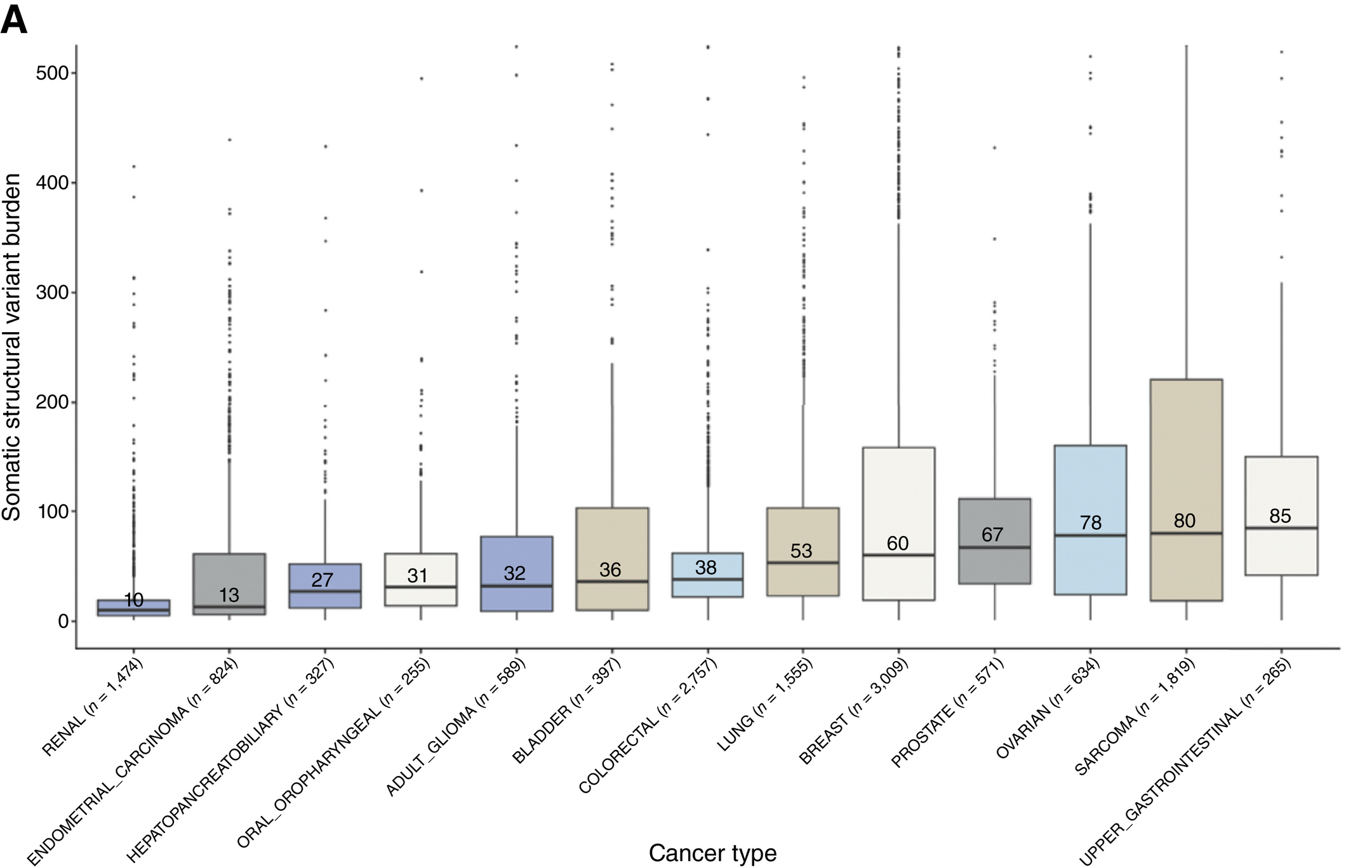
Mostly because high throughput sequencers are really good at detecting these, but we know that other types of variants, like structural variants (SVs - where big chunks of the genome have rearranged themselves), are just as important in cancer!
In the figure above, you can see that all of the major cancer types contain SVs and the nice thing about SVs is that there’s evidence that they’re some of the earliest mutations to develop during oncogenesis (when normal cells turn into cancer cells).
This means that they’re also probably going to stick around and not get lost as a tumor evolves.
But SVs also offer another important advantage:
They’re REALLY easy to detect using PCR!
And using PCR for MRD detection has distinct advantages over sequencing because it’s a much faster process but also, since it has a lower background, PCR can be 2-3 log more sensitive than sequencing.
What that means for patients is that we can catch recurrences sooner, reduce false negatives, and intervene before symptoms return or cancer spreads again!
###
Figure: Elliot MJ, et al. 2025. Clin Can Res. DOI: 10.1158/1078-0432.CCR-24-3472
Disclosure: I am the VP of Technology Development at SAGA Diagnostics - a company that uses SVs in MRD testing
The first 3D structure of DNA was published in 1938. It was generated by Florence Bell, a scientist you need to know.
One thing that we lose in the coverage of major scientific discoveries is all of the work that those advances were built on.
A perfect example of this is Watson and Crick's solving of the structure of DNA in 1953.
This wasn't just a synthesis of data they obtained from Maurice Wilkins via Rosalind Franklin (and others), but it also wouldn't have been possible without the early work of William Astbury and Florence Bell.
This is that story.
In the 1930's, William Astbury spent his days analyzing biological fibers using a technique called x-ray diffraction.
He was particularly interested in coiled fibers like those found in wool (keratin) and other textiles and was the first to characterize the structural changes in proteins as they are stretched.
He referred to these as the α-form and β-form, and laid the groundwork for Linus Pauling's discovery of the α-helix and β-strand in 1951!
Astbury quickly got a reputation for his expertise in x-ray diffraction, and was sent purified biological material to characterize from all over Europe.
In 1937, apparently elbows deep in things to characterize, he sent a letter to another renown crystallographer, Lawrence Bragg, asking if he knew of any rockstar crystallographers that were available to help with his studies.
Bragg recommended Florence Bell, a Cambridge grad who learned how to do crystallography from yet another x-ray luminary, JD Bernall.
A short correspondence later and Bell left her job working for Bragg to start her graduate work under the tutelage of Astbury.
While her research was focused on lots of different biological fibers, Bell's most important work was with DNA.
She showed that the key to obtaining the most informative images was the use of high molecular weight DNA.
The best source for this was calf thymus, and the diffractions she obtained from these fibers indicated that DNA had an ordered structure.
This finding was published in 1938 and was also presented at Cold Spring Harbor later that year by Astbury.
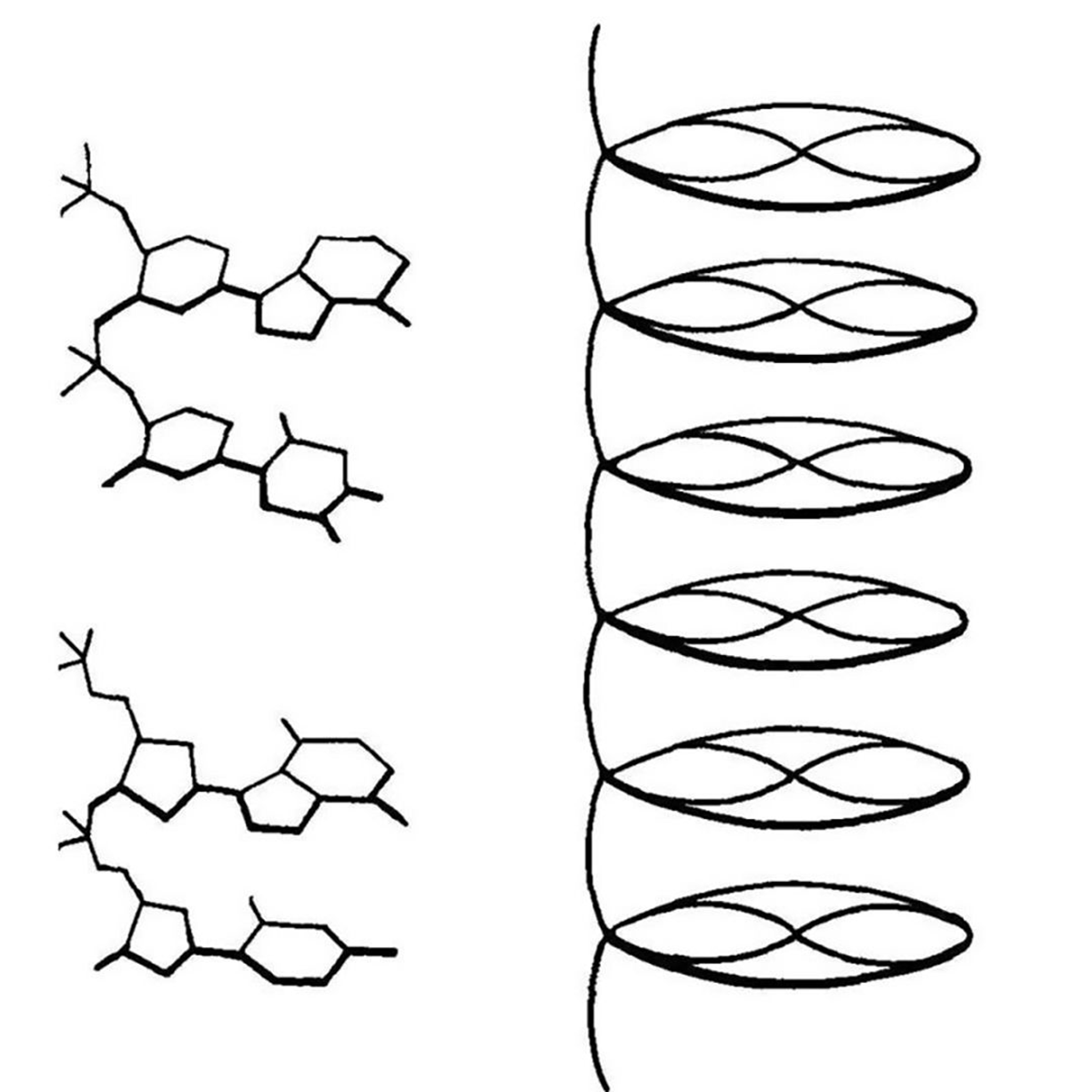
They described their structure of DNA as a 'pile of pennies' and a depiction of it can be seen in the figure above.
Their calculations also showed that the DNA nucleotides were 3.3 Å apart and they were stacked one on top of another.
While this might not seem like a historic achievement, it set the foundation for everything that would follow.
With a few tweaks and the discovery that DNA can exist in two forms depending on the humidity during drying, Rosalind Franklin and Raymond Gosling built on Bell's early work to generate photo 51 - the key diffraction of DNA used to solve the structure of DNA in 1953.
###
Astbury WT, Bell FO. 1938. Some recent developments in the x-ray study of proteins and related structures. CSHL Symp Quant Biol. DOI: 10.1101/SQB.1938.006.01.013
Weekly Reading List













Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here: