Omic.ly Weekly 80
June 23, 2025
Hey There!
Thanks for spending part of your week with Omic.ly!
This Week's Headlines
1) Loss of Y could be helping to make tumors in men more aggressive
2) Amino acids? We don't need more stinking amino acids...or do we?
3) Linus Pauling got the 3D structure of DNA wrong
4) Weekly Reading List
Who needs a Y chromosome anyway? Losing it could be making tumors in men more aggressive

Loss of the Y chromosome (LOY) is one of the most common somatic mutations to occur in men as they age.
Its loss has been shown to be associated with increased risk for cancer and poorer prognosis in those diagnosed with cancer.
However, it wasn't known if there were actual biological consequences for LOY or if this was just a passenger mutation that had no functional role in cancer risk or tumor progression.
LOY is often seen in Peripheral blood mononuclear cells (PBMCs) which are immune cells circulating in the blood.
PBMCs include key players like T cells, B cells, natural killer cells, monocytes, and dendritic cells which raises the possibility that LOY in these cells could compromise immune surveillance.
Immune surveillance is obviously important for detecting and fighting infections but its also super important in the development and progression of cancer!
It's known that the immune system plays a key role in identifying and destroying early cancer cells, so it begs the question whether LOY as men age contributes to deficiencies in the immune system that allow cancer cells to thrive.
That was the question posed recently by researchers who looked at LOY across multiple cell types and showed a distinct correlation between LOY and cancer risk which can be seen in the figure above:
a) heat-map of genes expressed across cancer types grouped by cells with and without LOY
b) LOY is common across cancer
c) the researchers developed a transcriptional score to predict LOY (YchrS) with an AUC of 98%
d, e, g) LOY increases with age, is similar across ethnicities, and is negatively correlated with cancer-testis antigen (CTA)
h) LOY is correlated with worse overall survival (OS) and worse disease-specific survival (DSS - death from cancer being studied)
The researchers went on to show that LOY in PBMCs correlates strongly with LOY in both tumor-infiltrating immune cells and malignant epithelial cells.
This suggests a possible mechanism linking LOY across diverse cell types!
They also showed that LOY in CD4+ and CD8+ T cells leads to immune dysfunction with CD4+ T cells having a marked increase in regulatory T cell (Treg) signatures, promoting an immunosuppressive environment with reduced activation, signaling, and cytotoxic capacity, effectively impairing the immune system’s ability to fight tumors.
Clinically, patients whose tumors contained both LOY epithelial cells and LOY T cells had significantly worse outcomes than those with LOY limited to one cell type.
Further, the presence of LOY in T cells and epithelial tumor cells were found to be independent predictors of poor prognosis, highlighting the potential additive effect of LOY in accelerating cancer progression.
These findings are important because they provide a new view into cancer biology where evaluation of LOY could be used in the future for diagnostics, prognostics, and therapeutic engineering.
###
Xingyu C, et al. 2025. Concurrent loss of the Y chromosome in cancer and T cells impacts outcome. Nature. DOI: 10.1038/s41586-025-09071-2
Genetic code expansion might be the coolest thing in Omics that you've never heard of.

DNA codes for messenger RNA (mRNA) which is then translated into protein.
And it's proteins that perform the majority of the functions in our cells!
They can serve as structural components, as molecular motors delivering cargo to various regions of the cell, and they can function as enzymes performing chemistry that would otherwise be impossible.
Proteins are created by ribosomes which read mRNA and stitch together amino acids to form the final protein sequence.
They do this by binding to mRNA and transfer RNAs (tRNAs) that are loaded with specific amino acids.
The sequence of the amino acids in a protein is determined by the mRNA sequence (codon) binding to a complementary tRNA (anticodon).
Proteins are made using 20 common amino acids but there are over 500 of them found in nature!
Many of those extra amino acids are just modified common amino acids, but this presents an interesting challenge:
What if we could expand the number of amino acids that can be coded for by DNA??
Doing this would allow us to engineer totally new proteins!
And this is the focus of an exciting area of synthetic biology referred to as ‘genetic code expansion.’
We can get to these designer proteins through a number of different biological hacks.
One of them is to change what the natural base pairs (NBPs) code for!
There are 4 DNA bases (A, T, C, G), these are read 3 at a time to code for proteins and this ‘triplet code’ is referred to as a codon. There are 64 potential codons and 20 common amino acids. Each amino acid is coded for by approximately 3 different codons.
But, we have the tools to change that!
Stop Codons - Are triplet codes that don’t code for any amino acids but serve to tell a ribosome to stop; however, we can make tRNAs that actually bind to the stop codons and cause a ribosome to insert whatever amino acid is attached to that stop-codon-binding-tRNA!
4 base codons - We can create tRNAs that use 4 base codons instead of 3 base codons (but this can get tricky in a living organism!)
Codon/tRNA Reprogramming - We can engineer a cell to incorporate a different amino acid on a specific tRNA.
But, hacking what the natural base pairs code can have unintended consequences in the proteins we need to keep cells alive!
Thankfully, synthetic biologists have also been working to add unnatural base pairs (UBPs) to these systems and have been successful in creating organisms that can use 6! base pairs (A, T, C, G, X, and Y).
This means we can create our funky new proteins without affecting how other critical proteins are made, because we can make tRNAs that complement new triplet codes and recognize our unnatural X and Y bases.
Ultimately, this allows us to create new proteins that have desirable functions.
Genetic code expansion has important applications in academic research, protein and enzyme engineering, and therapeutic development.
The kaleidoscopic image below is the triple helix Linus Pauling proposed as the structure of DNA in February 1953. Here's why he got it so wrong:
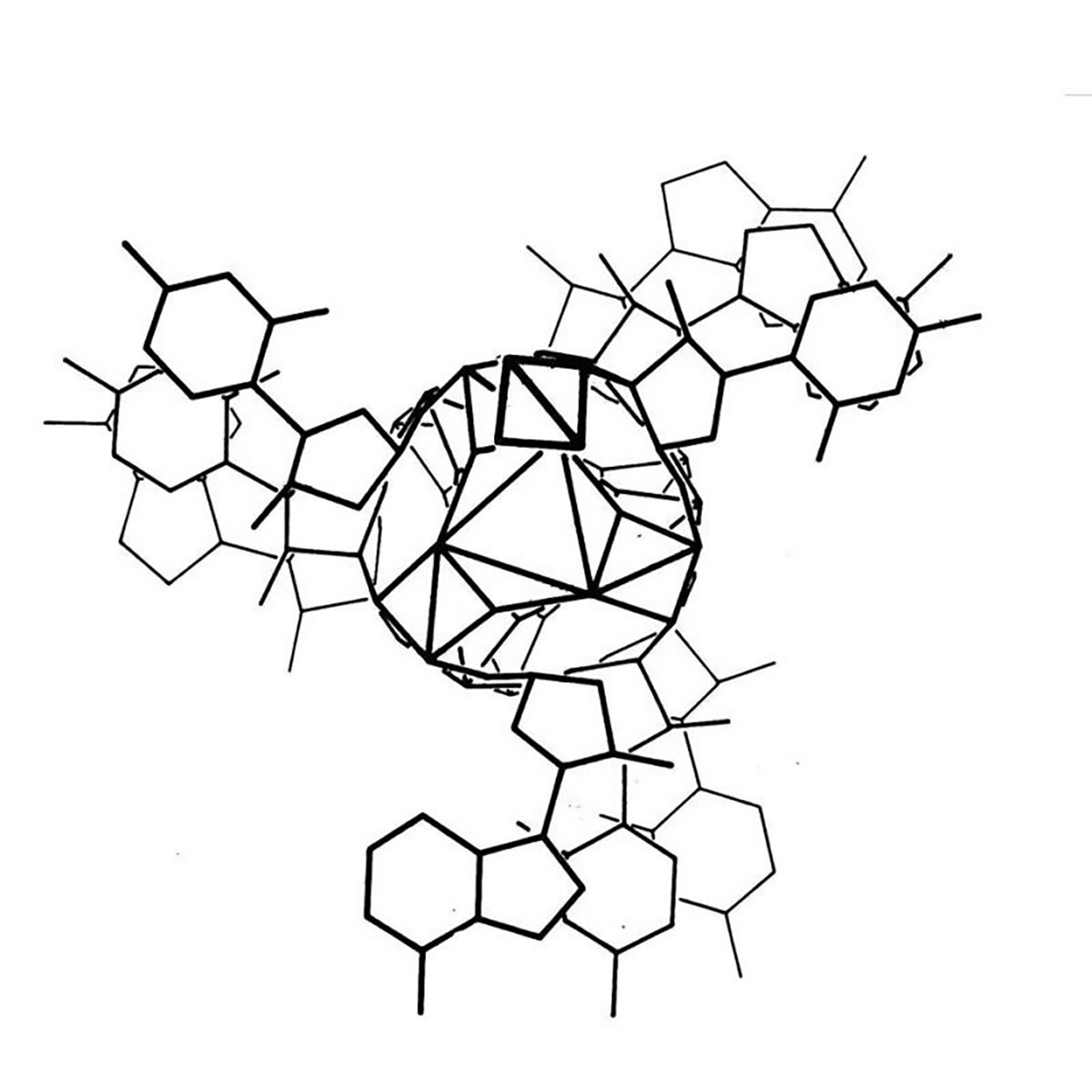
Linus Pauling is remembered as one of the greatest American scientists of all time. He was awarded two undivided Nobel Prizes, and is basically the father of modern structural biology.
He's most famous for solving the 3D structure of the α-helix and the β-sheet, the two most common protein structures.
To do this, Pauling employed a technique called x-ray crystallography which basically bombards a crystal with x-rays. How those x-rays bounce off of a crystal tells you something about the shape of the item that was crystallized and the resulting 'diffraction patterns' can be used to determine key structural features of the crystallized molecules.
In his work, Pauling took this a step further and was one of the first scientists to use that information to model molecular structures with balls and sticks.
Fresh off of his foundational work with proteins, Pauling turned his attention to DNA. While Pauling famously thought that DNA was NOT the genetic material, there's a rumor that he heard that teams in the UK were close to solving DNA's structure and the ever competitive Pauling wanted to see if he could beat them.
However, Pauling didn't generate his own data and relied heavily on data and calculations derived by others in the field, namely those created by William Astbury and Florence Bell in 1938.
Unfortunately, Pauling was unaware of the most recent advancements in the diffraction of DNA by Franklin and Gosling.
But he did have access to electron micrographs of DNA and the 15 year old Astbury/Bell data which indicated the structure was helical.
Based on incorrect density calculations, he also proposed that the structure had 3 nucleotides at each position.
It was already known that DNA was composed of a sugar phosphate backbone, so all of these things taken together led Pauling to a triple helix.
In the figure above, Pauling and Robert Corey placed the phosphate backbones of 3 DNA strands at the center of the structure with the nucleotides facing out, like the spokes of a bicycle wheel.
In hindsight, we know this is totally wrong and there are 3 key errors that make this structure implausible:
1. At pH 7, the phosphate backbone is negatively charged so 3 of them packed together would repel one another
2. The model doesn't leave space for sodium and Astbury's diffractions were all sodium salts
3. Chargaff's nucleotide pairing rules were totally ignored
Fortunately, taking inspiration from Pauling and his use of modeling, Watson and Crick, armed with pristine x-ray diffraction data from Franklin and Gosling, published the correct double helical structure for DNA two months later in the April 1953 issue of Nature.
###
Pauling L, Corey RB. 1953. A Proposed Structure For The Nucleic Acids. PNAS. DOI: 10.1073/pnas.39.2.84
Weekly Reading List









r/biotech Salary and Company Survey - 2025
by u/wvic in biotech





Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:






















