Sequencing exomes in healthy populations pays dividends
In a development that should surprise no one, population scale exome screening finds actionable variants.
When we completed the human genome project in 2003 it meant that genomics was basically over and scientists who did genomics had to pivot their careers to something else.
Unfortunately, that’s what happens when you complete things.
Kidding.
The Human Genome was only ~90% complete in 2003 (most of the coding genes were covered), and a gapless assembly of it wasn’t finished for another 19 years.
But even knowing where all of the genes in the human genome were located wasn’t super helpful.
We still had to figure out what they all did and how they contributed to health and disease.
And along the way, we realized that genetics is WAY more complicated than we thought!
This has meant that there’s been pretty good job security in genomics, because there’s still a lot of work to do to understand what’s going on here!
But that doesn’t mean we haven’t learned anything: we’ve been able to translate a bunch of those learnings into clinical practice.
We’ve identified hundreds of thousands of variants that lead to disease and are able to use genetic tools to detect these variants in people afflicted with genetic diseases.
Much of this testing, though, is reactive and relies on us seeing a phenotype or knowing that a patient has a family history of disease before we sequence them.
This is a very conservative approach to genetic testing and it means we miss a lot of diseases in people who don’t have (or don’t know they have!) a family history of disease or who are carriers of disease mutations but just aren’t showing symptoms yet!
What if we took a much less conservative approach and just sequenced anyone that wanted to be sequenced?
This might sound like a crazy idea, but sequencing is now cheap enough for us to do this at scale.
And at least one hospital system felt the same way!
Back in 2007, Geisinger health launched the MyCode project to biobank patient samples and link them to their EHR for future genetic analyses.
In 2014, they started a collaboration with the pharmaceutical company Regeneron to exome sequence all of the participants and return any Pathogenic/Likely Pathogenic (P/LP) results that were discovered in the sequencing process.
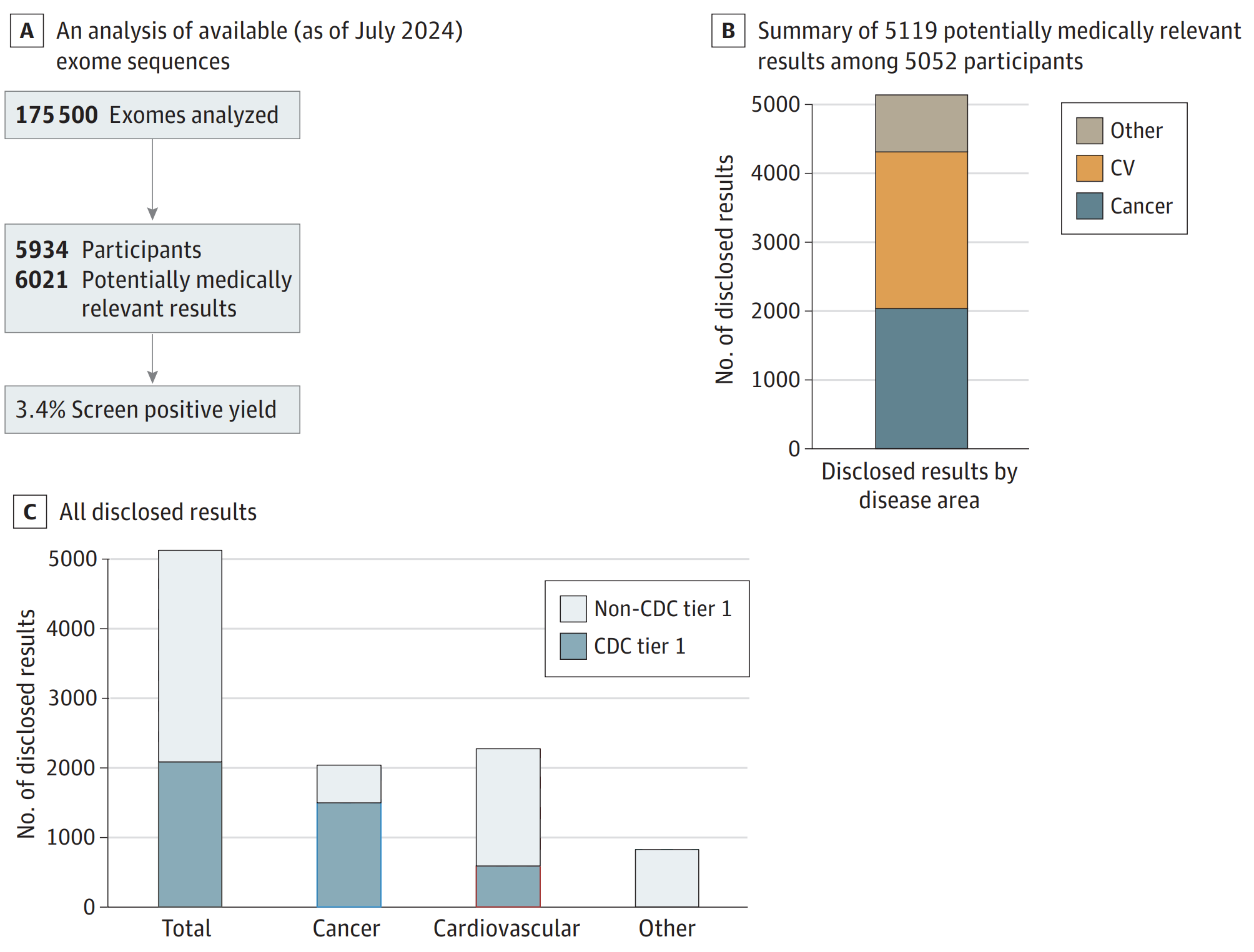
The results of this work can be seen in the figure above where A) 175,000 exomes have been sequenced and analyzed with a P/LP yield for 81 “actionable” genes of 3.4% B) of those results, the majority were related to risks for cardiovascular disease (CV) and Cancer and C) with about 1/3rd falling in CDC tier 1 targets (Hereditary Breast and Ovarian Cancer Syndrome, Lynch syndrome, and Familial hypercholesterolemia).
They also showed that ~90% of the people with a risk for developing disease were unaware they carried a disease causal mutation.
Now, you might be thinking that a yield of 3.4% might not sound like a lot, and if exomes now cost about $100 each to do this sort of analysis, it might not be worth it!
But when you consider we spend about $500B a year on cancer and cardiovascular disease treatments, exome sequencing everyone in the US (one time) for $300B might not be that bad of a deal after all.
And that’s especially true if patients, as they did in this study, make changes to help mitigate the risks of the pathogenic variants hiding in their genomes.
