STAMP brings us non-destructive single-cell profiling without the sequencing
No sequencer needed: Imaging based single-cell multiomics
Single-cell transcriptomic profiling has emerged in recent years as a powerful tool for understanding the biology of complex cellular mixtures and tissues.
It's been used extensively to look at how cells respond to changing environments, tease apart the dynamics of disease states, and track cell fates during development.
And looking at single-cells is important because the activities of cell populations or rare individual cells can get washed out in bulk measurements of cellular material.
So, understanding the role that individual cells play in development, the function of a tissue, or even the development of cancer can be really important!
Unfortunately, one of the biggest challenges facing the field of single-cell transcriptomics is that it's very expensive and requires a ton of sequencing to get useful information out of each cell.
The methods for isolating cells can also be technically challenging, influence what genes are expressed or even bias the results against specific cell populations that are hard to isolate.
The go-to methods in single-cell fall into four broad categories:
Flow Assisted Cell Sorting - A cell sorter is used to separate single cells and spit them out individually into a microtiter plate
Droplet-based Microfluidics - single cells are encapsulated in an oil droplet with reagents that lyse the cell and barcode the contents for sequencing
Microwells - Cells are distributed across wells of a microplate with the hope that each well only ends up getting a single cell
Combinatorial Barcoding - Cells are combined across multiple plates each containing unique barcodes - new barcodes are ligated after each move which results in each cell ending up with a unique barcode!
But what ALL of these methods have in common is that they end up destroying the cells and turning them into sequencing libraries.
This means those cells can't be used for any other downstream analyses like proteomic profiling.
It also means you're probably going to spend a lot of money sequencing these things!
But as single-cell studies have increased in popularity, so have spatial studies!
These two things are related but different, in single-cell you're isolating and looking at...single-cells - in spatial, you're usually looking at where transcripts and proteins are located in tissue sections on slides.
Most of the latest generation of spatial techniques use imaging to locate transcripts and antibody bound proteins which makes them a bit cheaper because they don't require sequencing AND they have the added benefit that the tissue sections are not destroyed during the different analyses.
Now, some smart people have realized that you can use these spatial imaging systems to also look at single-cells!
They've developed something called Single-Cell Transcriptomics Analysis and Multimodal Profiling (STAMP) and basically what's done here is that single-cell suspensions are fixed and permeabilized using formaldehyde, bound to a microscope slide, and then processed like they're any old tissue sample going into the spatial imaging process!
It's one of those things that's so simple and cool it makes you wonder why it took people so long to figure out this was possible!
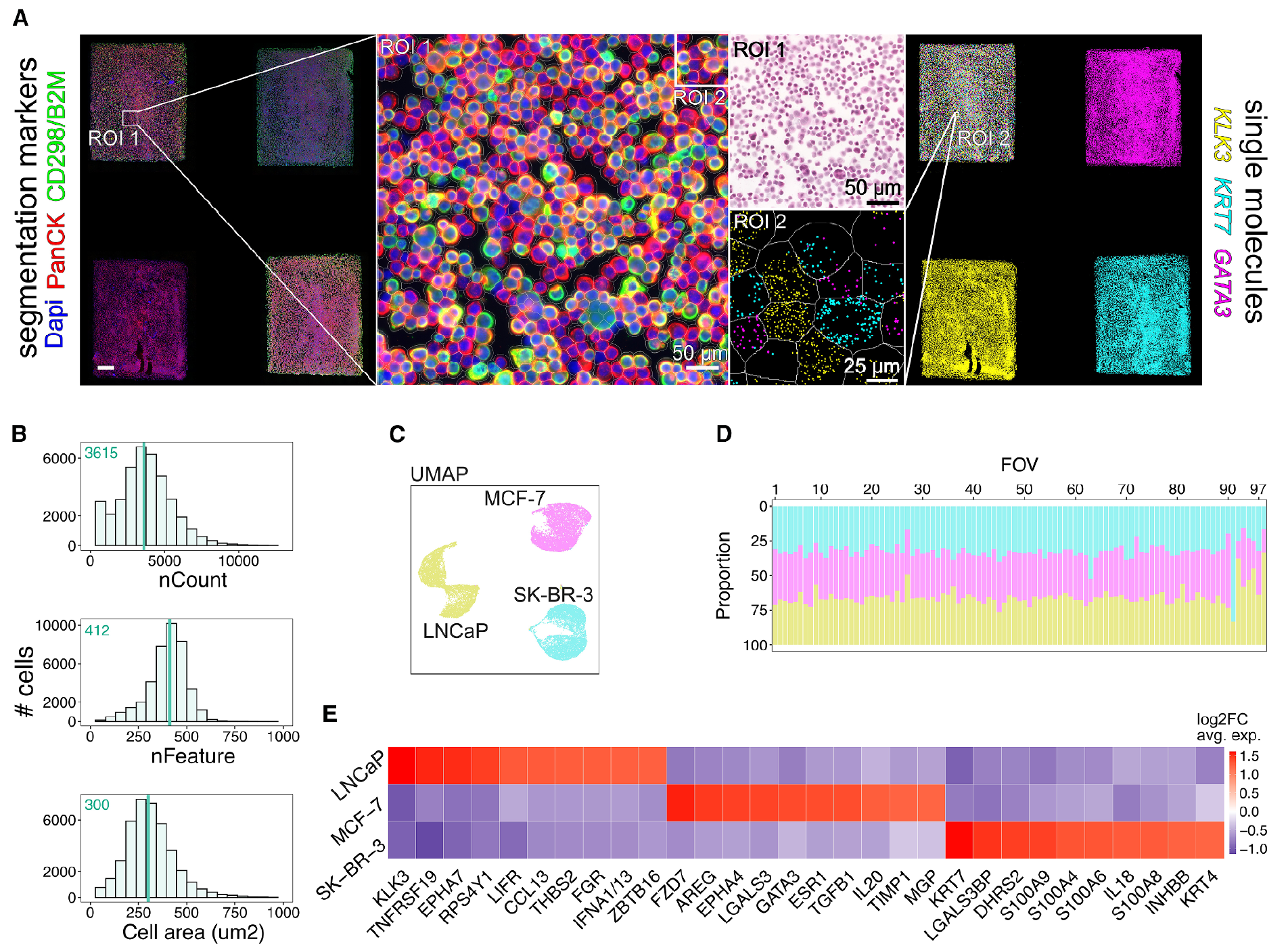
The results of this work can be seen in the figure above where a STAMP experiment was carried out with a 3-way mixture of MCF-7, LNCaP, and SK-BR-3 cancer cells on the CosMx platform.
A) shows the cell segmentation and transcript detection analysis, B) highlights that they get a lot of data from a lot of cells, and C,D,E) show they're able to easily discriminate the cells within the mixture!
They go on to show that they can detect a single MCF-7 cell in mixtures of millions of cells (1:100,000, 1:50,000) which highlights the impressive sensitivity of the technique for profiling things like circulating tumor cells in blood samples.
But the coolness doesn't stop there, because this technique is non-destructive so you can also do spatial proteomic profiling or analyze the samples across multiple different instruments.
The authors estimate that this technique will end up costing about $0.003 per cell which is kind of mindblowing when the average cost for a traditional sequencing based single-cell experiment is in the $0.10 range.
Now, there are drawbacks such as this technique doesn't produce full length transcripts so if you're into that you'll have to stick with the sequencing based techniques!
But STAMP looks like an interesting (and cheap) option for doing single-cell proteo-transcriptomics on a budget!
