Will Ultima's ultra cheap bases be a major boon to MRD?
Ultima Genomics is put to the test in MRD
Ultima Genomics came out of stealth in 2022 claiming they were going to upend the sequencing market with a very high throughput instrument that uses silicon wafers to perform the sequencing reaction.
The benefit of Ultima's spinning wafer scheme is that reagents can be flowed over the entire surface of the wafer allowing for very efficient distribution of reagents which helps to decrease costs.
Early reviews of Ultima's UG100 sequencer were mixed because it struggled to sequence homopolymers and the quality was just ever so slightly worse than Illumina.
But they seem to have fixed those problems and now offer a very high throughput system with a cost per base of about half that of its competitors.
This is important because one of the biggest roadblocks to using whole genome sequencing for cancer detection in liquid biopsies has been cost.
A liquid biopsy is just a blood based test that looks for the hallmarks of cancer in the blood.
This is done by sequencing the DNA floating around in the sample to see if we can find fragments with mutations in them.
These mutations can be indicative that someone has cancer, but these fragments are generally very rare, so you have to do A LOT of sequencing to find them!
We've come up with some clever ways to combine tumor informed sequencing (sequence a tumor to find important variants) with targeted capture of tumor mutations to reduce the amount of sequencing we need to do to detect cancer in liquid biopsies.
But it'd be much easier if we could take a blood sample and just sequence it without having to do a bunch of stuff up front!
Since cost is less of an issue on the Ultima platform, researchers at the New York Genome Center thought it might be perfect for performing tumor naive (where you don't sequence the tumor!) minimal residual disease testing (MRD).
MRD is a type of liquid biopsy where you're trying to determine how much cancer is in the body before and after it is treated to monitor when it might recur.
Since detecting recurrence early can have a positive impact on outcome, it's important to detect it as soon as possible which typically means more sequencing is better.
It's a great use case for Ultima's cheap bases!
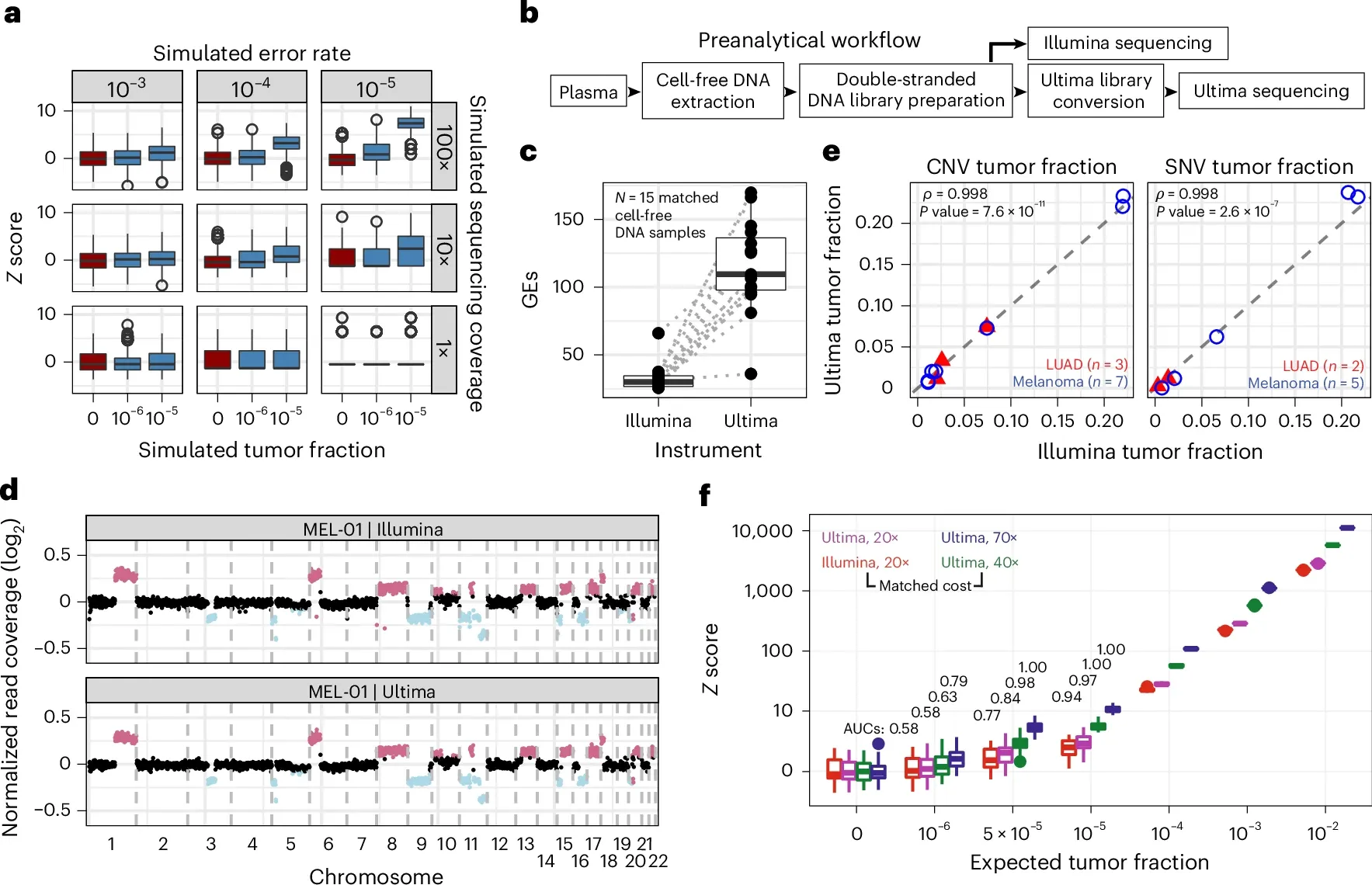
But before the researchers used the Ultima sequencer for MRD testing, they compared its performance to that of Illumina which can be seen in the figure above. a) is a comparison of sequencing error rates to tumor mutations and shows that to detect tumor fractions below 10-5, you need at least 100x coverage and an error rate of less than 10-4. b) is a workflow diagram showing how they performed the comparison. c) displays a graph of sequencing coverage for each sample. d,e, and f) show that the results from Illumina and Ultima are almost identical with the higher coverage on the Ultima samples (f) showing greater sensitivity for lower frequency variants!
The researchers go on to show that using duplex error corrected reads (ppmSeq) that they're able to use whole genome MRD to track recurrence in a mouse model of melanoma and in patients with urothelial cancer.
This work is clinically important because it's not always possible to get an informative chunk of tumor for performing MRD testing.
In those scenarios, deeply sequencing a patient's blood on a UG100 might be the next best option!
