The genetics of pregnancy loss
10-20% of pregnancies end in miscarriage. Genetics is helping us to explain why that is!
Pregnancy loss can be devastating to families and we’ve known for a while that there are multiple factors that can contribute to the loss of a fetus.
These can include maternal infection, chronic disease (diabetes, high blood pressure, obesity), structural abnormalities in the uterus, and hormonal problems.
But one of the biggest contributors to pregnancy loss is genetics.
We know that chromosomal aneuploidies (where extra chromosomes a present) can lead to miscarriages; however, the contribution of other genetic abnormalities to pregnancy loss is less well understood.
These abnormalities can include novel combinations of small sequence variants carried by each parent that lead to disease, non-disjunction of chromosomes (chromosomes don’t separate during meiosis), de novo mutations (mutations not carried by either parent) that occur during the formation of gametes (sperm and eggs), or even errors that occur during chromosome recombination (deletions, duplications, translocations, and inversions).
Recombination itself is an important evolutionary adaptation that allows for the mixing of a genome to create the diversity required for a species to survive in a changing environment.
This is much less important now as we’ve developed into an advanced species with things like houses and healthcare, but understanding how genetic changes continue to impact our health and ability to reproduce has major societal impacts.
Studying these changes was the focus of the Copenhagen Pregnancy Loss (COPL) Study which recently published their results in Nature.
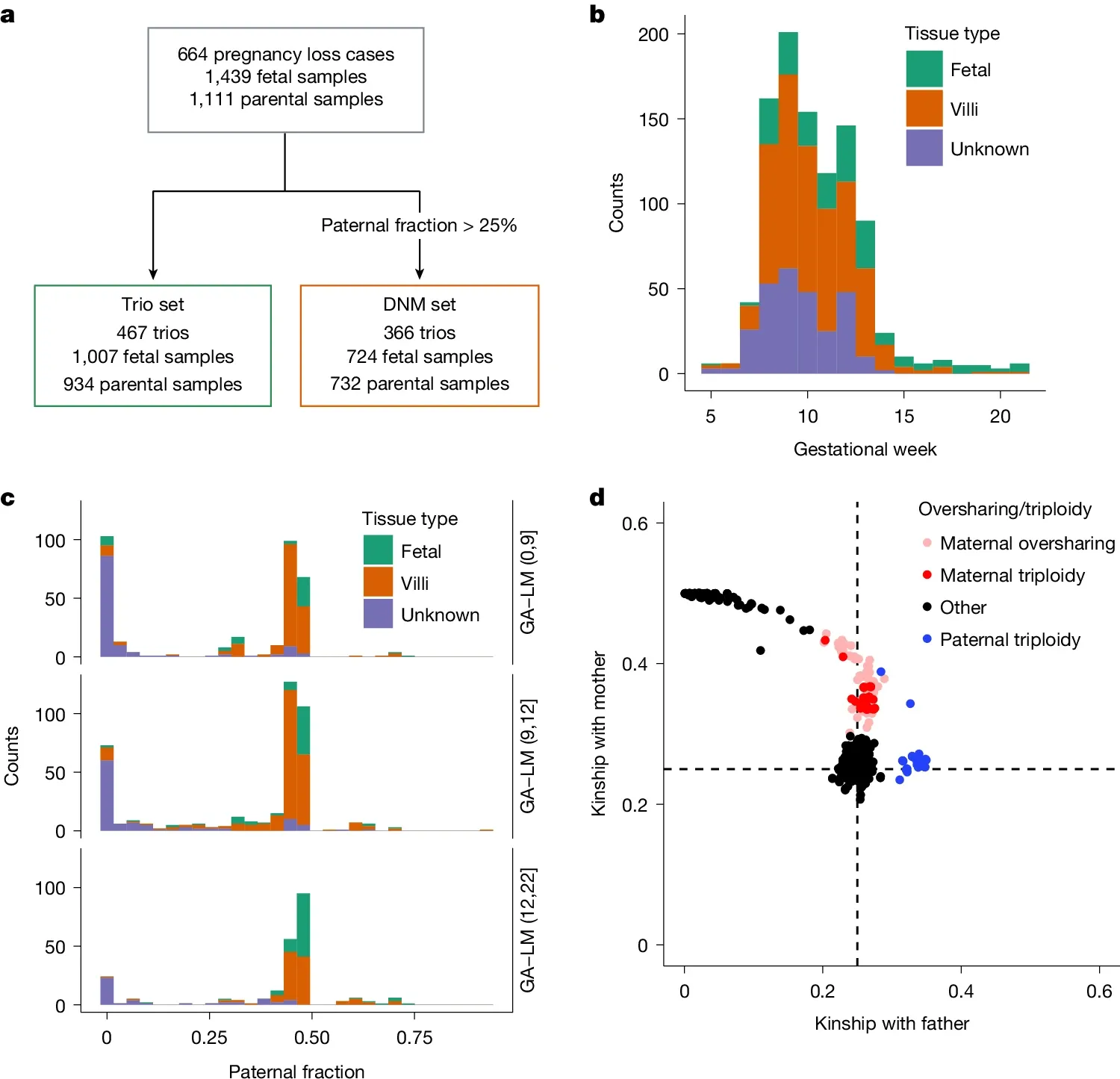
They performed short-read whole genome sequencing on 467 trios (fetus, mother, father) to identify genetic abnormalities that might explain early embryonic demise.
The figure above is an overview of the study and sample set. a) is a flowchart of the study, b) shows the gestational age of the samples before loss, c) is the paternal fraction observed (a measure of maternal cell contamination), indicating that this sample set showed maternal cell contamination and d) is a similar plot showing relatedness to the father highlighting a skew toward maternal genetics (a combination of contamination and the fact that some of these samples were maternal triploidies - 2 copies of mom + 1 copy of dad).
This study found that 55% of pregnancy loss cases could be explained genetically:
44.1% by aneuploidy
6.4% by triploidy
3.3% by pathogenic small sequence variants
1.3% by de novo copy number variants
They also saw that the maternal genome contributed most frequently to these abnormalities, which is consistent with meiotic errors during oogenesis.
However, euploid (the correct number of chromosomes) fetuses without structural anomalies still showed a significant burden of pathogenic mutations.
Further, the study estimated that 1 in 136 pregnancies could be lost due to pathogenic small sequence variants, underscoring the need to look beyond chromosomal abnormalities in pregnancy loss cases.
